LiQA
Liver Fibrosis Quantification and Analysis
Motivation
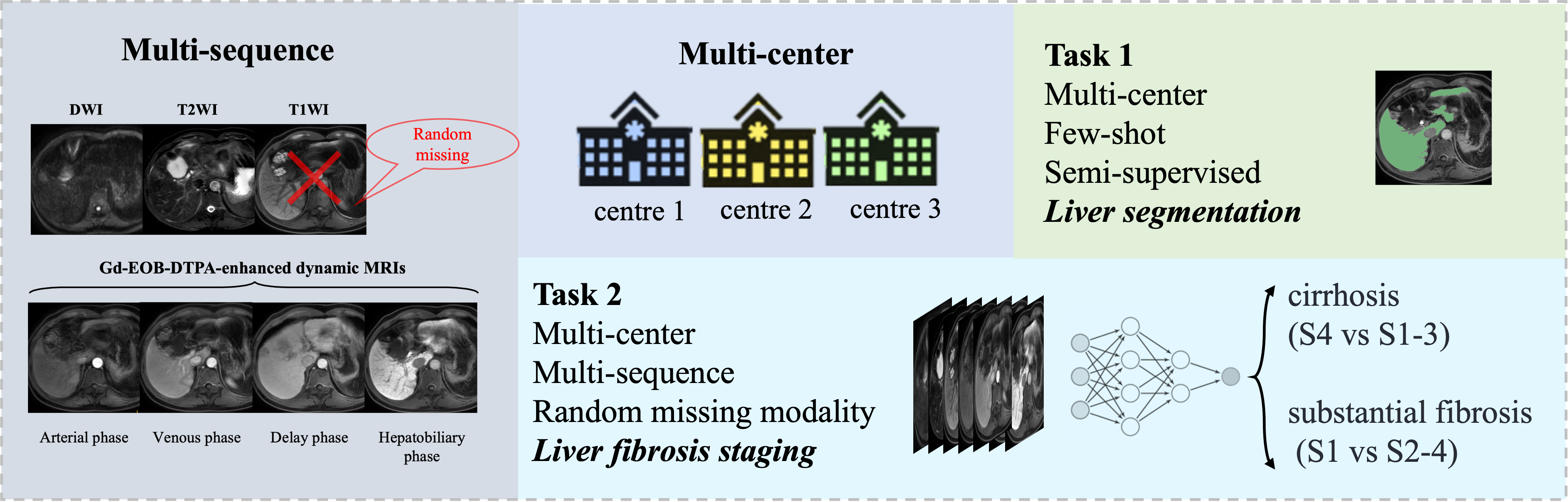
Liver fibrosis, arising from chronic viral or metabolic liver conditions, presents a significant global health challenge. Accurate liver segmentation (LiSeg) and fibrosis staging (LiFS)
Task
Task 1: The LiSeg task aims to predict liver segmentation with limited ground truth, where the Hepatobiliary phase (HBP) MRI is provided, which contains crucial information on the liver.
Task 2: The LiFS task aims to stage liver fibrosis accurately. The severity of liver fibrosis can be classified into four stages (S1-S4). Results of two binary classification tasks with clinical significance are evaluated, i.e., staging cirrhosis (S1-3 vs S4) and identifying substantial fibrosis (S1 vs S2-4).
Notably, both LiSeg and LiFS encounter domain shifts across multi-center data. Participants are encouraged to integrate complementary information from multi-phase MRIs effectively, achieving more precise and generalizable results. Moreover, automatic LiFS may face challenges including random missing sequences for certain patients and misalignments among multi-phase MRIs.
To address this task, participants are also encouraged to leverage external data, such as LLD-MMRI2023, and pretrained models.
Data
About the data
1) Scanner: Philips Ingenia3.0T, Siemens Skyra 3.0T, Siemens Aera 1.5T.
2) Dataset overview: The track cohort was composed of 440 patients diagnosed with liver fibrosis who underwent multi-phase MRI scans. All subjects were scanned in clinical centers using three different magnetic resonance scanner vendors. The dataset will include multi-phase and multi-center data, consisting of T2-weighted imaging, diffusion-weighted imaging, and Gadolinium ethoxybenzyl diethylenetriamine pentaacetic acid (Gd-EOB-DTPA)-enhanced dynamic MRIs. Gd-EOB-DTPA-enhanced dynamic MRIs include the non-contrast phase (T1WI), arterial phase, venous phase, delay phase, and hepatobiliary phase.
3) Contrast-enhanced dynamic scans: Contrast-enhanced scans were performed based on the injection of the GD-EOB-DTPA agent. The arterial phase is captured 25 seconds after the contrast agent is injected. Subsequently, the portal phase is achieved 1 minute later. After another 3 minutes, the delay phase is obtained, and finally, the hepatobiliary phase is reached 20 minutes thereafter.
4) Data format: The data are all in Nifty format. Each sample may randomly lack phases (except hepatobiliary phase ), and the sequences have not applied pre-alignment through spatial registration.
Training Set
For LiSeg task, the training set contains 30 annotated segmentation images obtained by experienced clinicians and 220 unannotated images from 2 different MRI vendors and 3 different centers.
For LiFS task, all 250 training samples are provided with the ground truth of the liver fibrosis stage. Moreover, part of the hepatobiliary phase images have been segmented previously.
| Vendor | Center | Num. studies | Num. Annotations |
|---|---|---|---|
| A | A | 100 | 10 |
| B | B1 | 100 | 10 |
| B | B2 | 50 | 10 |
Validation Set
| Vendor | Center | Num. studies | Num. Annotations |
|---|---|---|---|
| A | A | 10 | 10 |
| B | B1 | 10 | 10 |
| B | B2 | 10 | 10 |
Test Set
The 160 test cases corresponded to 120 new cases from the vendors provided in the training set and 40 additional cases from a third unseen vendor, that were tested for model generalizability.
| Vendor | Center | Num. studies |
|---|---|---|
| A | A | 40 |
| B | B1 | 40 |
| B | B2 | 40 |
| C (new) | C | 40 |
Metrics & Ranking
Metrics
- LiSeg: Dice Similarity Coefficient (DSC), Hausdorff Distance
- LiFS: Area under curve (AUC), Accuracy;
Rank methods
The ranks of classification and segmentation tasks are ranged separately.
- a. For classification, the average of metrics, i.e., Area under curve (AUC) and accuracy, are utilized to evaluate performance.
- b. For segmentation, the average of metrics, i.e., Dice Similarity Coefficient (DSC) and Hausdorff Distance is utilized to evaluate performance.
Finally, the average of in-distribution results (seen center) and out-of-distribution results (unseen center) are used for the final ranking.
Rules
- Publicly available data (such as LLD-MMRI2023) and pre-trained models are allowed.
- Only automatic methods are acceptable.
Registration
To access the dataset, please register in the LiQA registration platform.
Submission Guidance
Model Submission
After registration, we will assign participants an account to login into our LiQA evaluation platform. Participants can directly upload your predictions on the validation data (in nifty format) via the website. Note that evaluation of validation data will be allowed up to 10 times for each task per team. For fair comparison, the test dataset will remain unseen. Participants need to submit their docker models for testing.
Paper Submission
Please refer to our paper submission guidance.
Timeline
The schedule for this track is as follows. All deadlines(DDLs) are on 23:59 in Pacific Standard Time.
| Training Data Release | May 10, 2024 |
|---|---|
| Validation Phase | |
| Test Phase | |
| Abstract Submission | |
| Paper Submission | August 15, 2024 (DDL) |
| Notification | September 15, 2024 |
| Camera Ready | September 25, 2024 (DDL) |
| Workshop (Half-Day) | October 10, 2024 |
Citations
Please cite these papers when you use the data for publications:
@inproceedings{gao2023reliable,
title={A reliable and interpretable framework of multi-view learning for liver fibrosis staging},
author={Gao, Zheyao and Liu, Yuanye and Wu, Fuping and Shi, Nannan and Shi, Yuxin and Zhuang, Xiahai},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={178--188},
year={2023},
}
@misc{liu2024merit,
title={MERIT: Multi-view Evidential learning for Reliable and Interpretable liver fibrosis sTaging},
author={Yuanye Liu and Zheyao Gao and Nannan Shi and Fuping Wu and Yuxin Shi and Qingchao Chen and Xiahai Zhuang},
year={2024},
archivePrefix={arXiv},
}
Contact
If you have any questions regarding the LiQA track, please feel free to contact:
- Jiyao Liu: jiyaoliu.fudan@gmail.com
- Yuanye Liu: yuanyeliu@fudan.edu.cn